Plants produce many short-interfering RNAs (siRNAs) that are often generated from endogenous loci and repeats or by exogenous agents such as virus. These siRNAs are derived from aberrant double-stranded RNAs and require the activity of RNA-dependent RNA polymerases (RDRs). Biogenesis of siRNAs requires the functions of DCL2, DCL3, DCL4, RDR2, RDR6, and RNA polymerase IV. In some cases, endogenous siRNAs trigger epigenetic effects on target loci and are associated with RNA-directed DNA methylation and chromatin remodeling. RDR6 plays a role in silencing of transgenes, some viruses, and specific endogenous mRNAs that are targets of trans-acting siRNAs (ta-siRNAs). ta-siRNAs are formed by miRNA-guided cleavage within pre-ta-siRNA transcripts (phasing) and through DICER-LIKE processing of dsRNA that is dependent on RDR6/SDS3 activity. The phased ta-siRNAs direct the degradation of target mRNAs as do plant miRNAs. It is predicted that divergence of siRNAs and their targets between species mediate hybrid incompatibility in allopolyploids.
MicroRNAs (miRNAs) are 20-24 nucleotide RNA molecules that play essential roles in posttranscriptional regulation of target genes. tasiRNAs are formed by miRNA-guided cleavage products within primary tasiRNA transcripts, and the phased tasiRNAs direct the degradation or repression of target mRNAs as do plant miRNAs. Unlike siRNAs, many miRNA and tasiRNA loci are conserved in sequences, and unique miRNAs and tasiRNAs were commonly identified in the allotetraploids and their progenitors. In plants, miRNAs predominately mediate degradation of target mRNAs via perfect or near-perfect complementary sequences. MicroRNA targets include a large number of transcription factors, suggesting a role of miRNAs in the control of regulatory networks and cellular growth and development. Many miRNAs and their targets are conserved among plants or animals, whereas some are specific to a few plant or animal lineages. Conserved miRNAs do not necessarily exhibit the same expression levels or patterns in different species or at different stages within a species. MicroRNA loci in different species inherited from ancestral species may diverge in sequence and expression patterns (e.g., tissue-specificity), gain new expression patterns, or undergo gene loss, as a consequence of genetic and epigenetic changes during evolution. miRNAs and expression of targets tend to be co-regulated, which suggests that miRNA biogenesis proteins such as DCLs, AGOs, and RDRs have concerted regulation within species and diversified modifications between species. Over time, progenitors’ miRNA loci may diverge expression and gain new expression patterns, as a consequence of genetic and epigenetic changes.
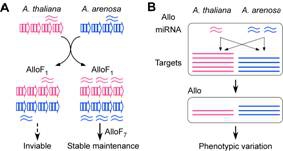
Combination of diverged progenitors’ loci in interspecific hybrids and new allopolyploids perturbs the regulatory balance in expressing miRNAs and their biogenesis genes. As a result, the miRNA accumulation levels are nonadditive, leading to nonadditive expression of their targets in interspecific hybrids and allotetraploids. This nonadditivity is partly associated with transcriptional regulation of miRNA loci and their targets. In addition, nonadditive expression of miRNA biogenesis genes and preference of miRNAs for repressing species-specific targets suggest a miRNA-dependent mechanism for repression of some progenitors’ genes observed in interspecific hybrids and allopolyploids. We are investigating how expression divergence between progenitors’ miRNA loci and their targets affect morphological evolution and phenotypic novelty in interspecific hybrids and allopolyploids.
Web information about small RNAs
The Nobel Prize in Physiology or Medicine (2006) on small RNAs
Non-coding RNAs
Small interfering RNAs
MicroRNAs
PiWi-interacting RNAs
Ten selected papers about small RNAs in related species and allopolyploids
[1-10]
- Axtell MJ, Bowman JL: Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343-9 (2008).
- Bourc'his D, Voinnet O: A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330: 617-22 (2010).
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC: Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-11. (1998).
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ: Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci U S A 106: 17835-40 (2009).
- Ha M, Pang M, Agarwal V, Chen ZJ: Interspecies regulation of microRNAs and their targets. Biochim Biophys Acta 1779: 735-742 (2008).
- Hamilton AJ, Baulcombe DC: A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950-2 (1999).
- Martienssen RA: Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol 186: 46-53 (2010).
- Napoli C, Lemieux C, Jorgensen R: Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279-289 (1990).
- Niwa R, Slack FJ: The evolution of animal microRNA function. Curr Opin Genet Dev 17: 145-50 (2007).
- Swaminathan K, Alabady MS, Varala K, De Paoli E, Ho I, Rokhsar DS, Arumuganathan AK, Ming R, Green PJ, Meyers BC, et al.: Genomic and small RNA sequencing of Miscanthus x giganteus shows the utility of sorghum as a reference genome sequence for Andropogoneae grasses. Genome Biol 11: R12 (2010).



